Hematological malignancies are characterized by multiple classes of chromosomal aberrations. Comprehensive detection of these aberrations is vital to properly classify samples and provide actionable information. Due to the limitations of current cytogenetic approaches, only a fraction of the samples evaluated lead to a positive aberration finding11-13. Optical genome mapping (OGM) significantly improves detection to unlock truly comprehensive chromosomal aberration profiles and maximize the number of informed cases.




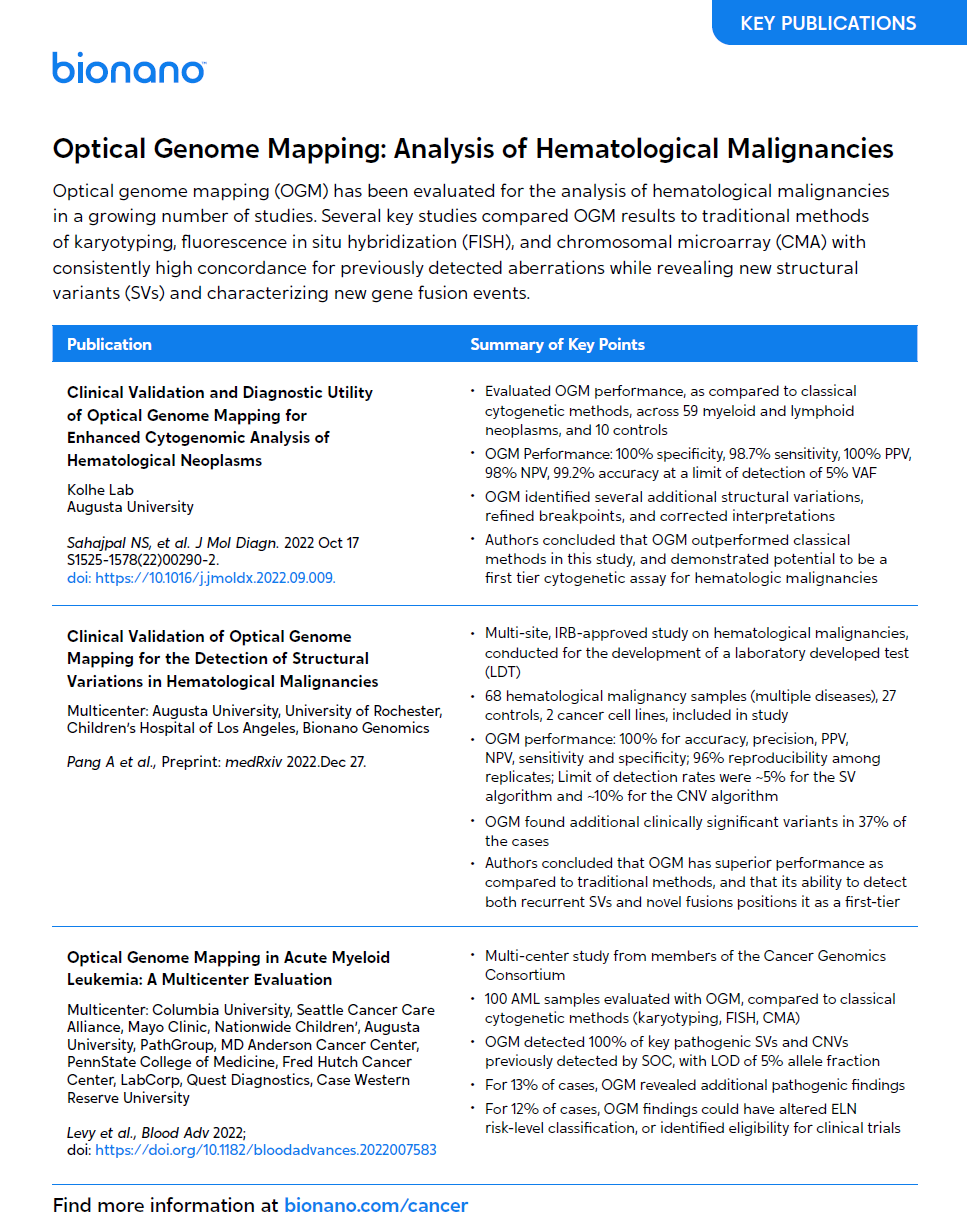
Multiple peer-reviewed studies from global experts have demonstrated the same outcomes: OGM has a high correlation to results from traditional methods across diverse types of hematological malignancies, while consistently revealing additional pathogenic findings.
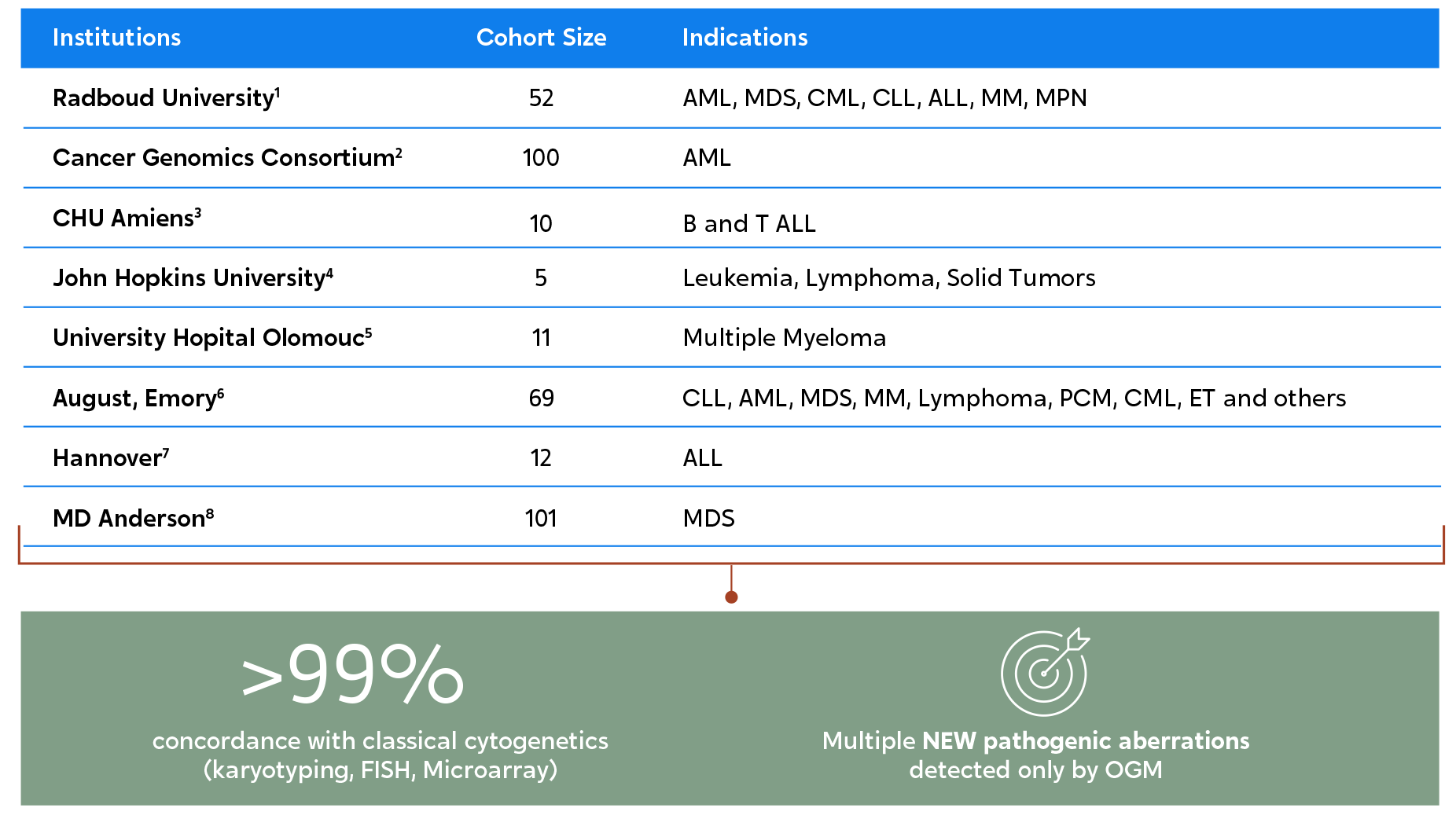
When compared to traditional cytogenetic methods, OGM can uncover up to twice as many pathogenic chromosomal aberrations.
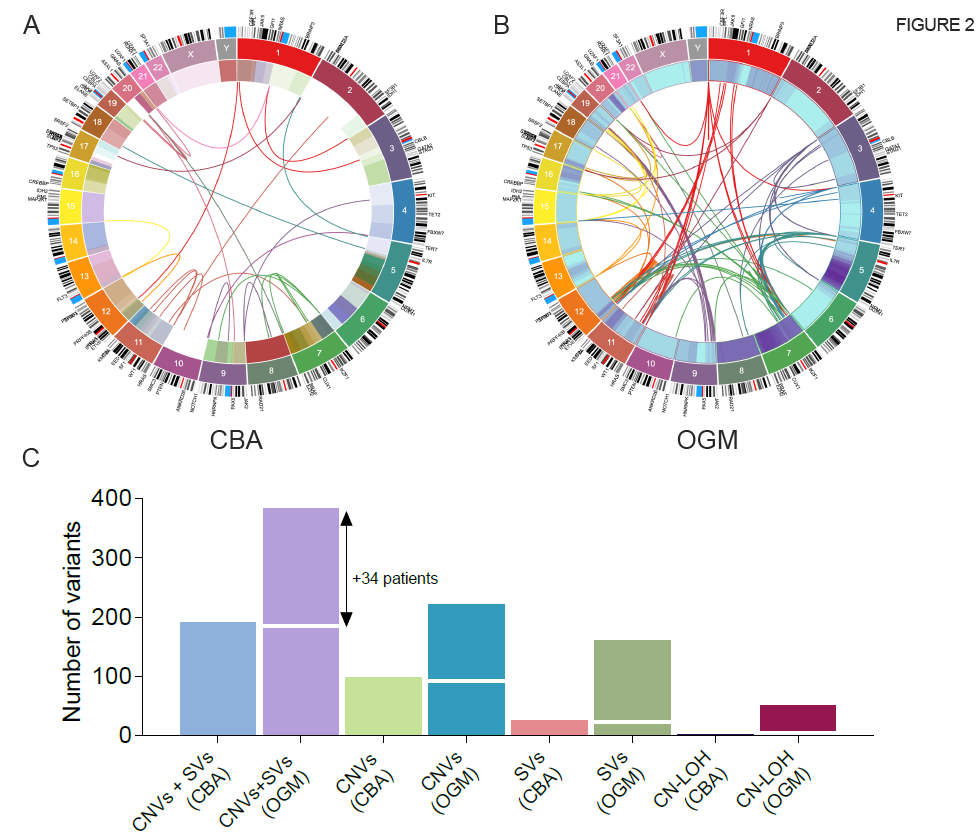
In a 2022 MD Anderson Cancer Center study, OGM detected twice the number of clinically significant chromosomal aberrations as karyotyping in 101 myelodysplastic samples.8 This figure illustrates the Circos plot view of the variants identified by karyotyping (left) versus OGM (right).
High-resolution and enhanced characterization of samples by OGM also leads to better risk analysis and stratification of samples.
| Disease | Impact of OGM Analysis |
| AML and MDS9 | For 67% of cases, the karyotype was redefined. In some cases, this led to an adjustment in ELN risk classifications |
| AML and MDS10 | 11% of samples for which OGM findings led to change in R-IPSS or 2010/2017 ELN score |
| MDS8 | 17% of cases assessed with OGM had the IPSS-R risk reclassified from initial karyotyping |
Sign up to watch the AMP 2022 Bionano Corporate Workshop #1, which discusses Maximizing Detection of Pathogenic Structural Variants Across Hematological Malignancies with Optical Genome Mapping.
Watch VideosSign up to watch this webinar and hear from Bionano’s Dr. Alka Chaubey and Dr. Adam Smith, UHN, as they discuss how Optical Genome Mapping Meets Challenges for New Blood Cancer Classifications
Watch On-demandSign up to watch this webinar and hear how OGM makes a huge impact on clinical research, as Dr. Rashmi Kanagal-Shamanna and Dr. Guillermo Garcia-Manero (MD Anderson Cancer Center) discuss OGM for Enhanced Characterization of Myelodysplastic Syndromes.
Watch On-demandRead about what structural variations are and why they matter.
Learn MoreSee how OGM reveals structural variation in a way that has never been done before.
Learn MoreFind the latest research in our Publications Library.
Learn More
| title | Source | Authors |
|---|---|---|
| Optical Genome Mapping Identifies Novel Recurrent Structural Alterations in Childhood ETV6::RUNX1+ and High Hyperdiploid Acute Lymphoblastic Leukemia | September 21, 2023 | Brandes, Danielle; Yasin, Layal; Nebral, Karin; Ebler, Jana; Schinnerl, Dagmar; Picard, Daniel; Bergmann, Anke; Alam, Jubayer; Köhrer, Stefan; Haas, Oskar A.; Attarbaschi, Andishe; Marschall, Tobias; Stanulla, Martin; Borkhardt, Arndt; Brozou, Triantafyllia; Fischer, Ute; Wagener, Rabea |
| Assembly of 43 diverse human Y chromosomes reveals extensive complexity and variation | June 12, 2023 | Pille Hallast, Peter Ebert, Mark Loftus, Feyza Yilmaz, Peter A. Audano, Glennis A. Logsdon, Marc Jan Bonder, Weichen Zhou, Wolfram Hoeps, Kwondo Kim, Chong Li, Philip C Dishuck, David Porubsky, Fotios Tsetsos, Jee Young Kwon, Qihui Zhu, Katherine M. Munson, Patrick Hasenfeld, William T. Harvey, Alexandra P. Lewis, Jennifer Kordosky, Kendra Hoekzema, The Human Genome Structural Variation Consortium (HGSVC), Jan O. Korbel, Chris Tyler-Smith, Evan E. Eichler, Xinghua Shi, Christine R Beck, Tobias Marschall, Miriam K. Konkel, Charles Lee |
| Babesia duncani multi-omics identifies virulence factors and drug targets | April 13, 2023 | Pallavi Singh, Stefano Lonardi, Qihua Liang, Pratap Vydyam, Eleonora Khabirova, Tiffany Fang, Shalev Gihaz, Jose Thekkiniath, Muhammad Munshi, Steven Abel, Loic Ciampossin, Gayani Batugedara, Mohit Gupta, Xueqing Maggie Lu, Todd Lenz, Sakshar Chakravarty, Emmanuel Cornillot, Yangyang Hu, Wenxiu Ma, Luis Miguel Gonzalez, Sergio Sánchez, Karel Estrada, Alejandro Sánchez-Flores, Estrella Montero, Omar S. Harb, Karine G. Le Roch, Choukri Ben Mamoun |
Optical genome mapping offers a groundbreaking method for studying the human genome with […]
For decades, the field of oncology has heavily relied on traditional investigative methods […]
One of the remarkable aspects of Optical Genome Mapping (OGM) is its ability […]
